Protocol
DNA oligo
We purchased the following DNA from Integrated DNA Technologies (IDT)
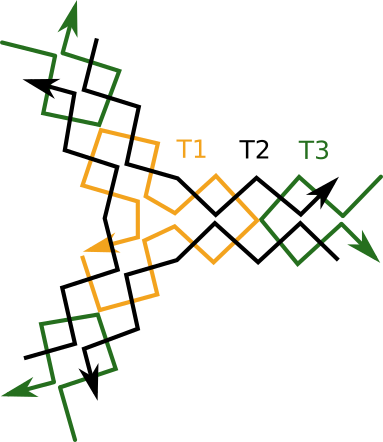
THREE POINT STAR (TPS)
| Name | sequence (5'→3') |
| T1 | aggcaccatcgtaggcttgccaggcaccatcgtaggcttgccaggcaccatcgatggcttgcc |
| T2 | actatgcaacctgcctggcaagcctacgatggacacggtaacg |
| T3 | tcgtgcgttaccgtgtggttgcatagtctgtt |
JOINT
| Name | sequence (5'→3') |
| J1 | cacgaacgcagcaagccacccaaccagaaagacacaaaacag |
| J2 | cacgaaaaagtctttctggttgggtggcttgctaaaaaacag |
| R | ctgttttttagcaagccacccaaccagaaagactttttcgtg |
Buffers and gels
TAE BUFFER
| Materials | Concentration |
| Tris-acetate (pH8.3) | 40mM |
| EDTA | 1mM |
TAE-MG BUFFER
| Materials | Concentration |
| Tris-acetate (pH8.3) | 40mM |
| EDTA | 1mM |
| Mg(OAc)2 | 10mM |
10%PAGE
| Materials | Quantity |
| water | 4.8ml |
| 40×TAE-Mg | 188ul |
| 30% m/v acrylamide | 2.5ml |
| TEMED | 7.5ul |
| 10% APS | 75ul |
15%PAGE
| Materials | Quantity |
| water | 3.55ml |
| 40×TAE-Mg | 188ul |
| 30% m/v acrylamide | 3.75ml |
| TEMED | 7.5ul |
| 10% APS | 75ul |
Electrophoresis
Samples were loaded on the PAGE gels described above (10% for TPS [three point star] unit, 15% for joints). TAE was used as running buffer. After 15V for 15 hour electrophoresis, PAGE gels were stained by Ethidium bromide. Stained DNA was observed by UV illuminator (365 nm).
The electrophoresis was conducted as below.
| | T1 | T2 | T3 | T1+T2 | T2+T3 | T3+T1 | T1+T2+T3 |
| T1(1uM) | + | | | + | | + | + |
| T2(3uM) | | + | | + | + | | + |
| T3(3uM) | | | + | | + | + | + |
| | J1 | J2 | J1+J2 |
| J1(2uM) | + | | + |
| J2(2uM) | | + | + |
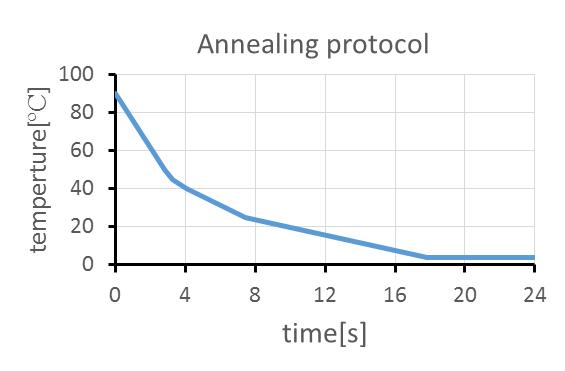
These DNA strands were hybridized by 90 ºC for 5 min, 90 ºC for 5 min, 65 ºC for 5 min, 50 ºC for 5 min, 37 ºC for 5 min,, and 25 ºC for 5 min. The DNA after annealing was stored at 4 ºC.
Under the following conditions DNA was occluded and then annealed to create the DNA tiles.
DNA concentrations are as follows.
| | TPS+dsDNA joint | TPS+dsDNA joint |
| T1 | 1uM | 1uM |
| T2 | 3uM | 3uM |
| T3 | 3uM | 3uM |
| J1 | 1.5uM | 1.5uM |
| J2 | 1.5uM | none |
For annealing, we conducted a temperature change program as the following protocol using ThermoGradient96 (BIOMETRA).
DNA tile sheets observation by Fluorescence microscopy
1/1000 dense MIDORI GREEN (Nippon Genetics) was mixed with newly-formed 5 uL of DNA tile before observation. DNA tiles were observed using a fluorescent microscope Axiovert 200M (Carl Zeiss, Jena, Germany) with a CMOS camera ORCA-Flash4.0 V2 (Hamamatsu Photonics, Shizuoka, Japan). A fluorescent filter for GFP (Zeiss) was used to detect DNA stained with MIDORI GREEN.
DNA tile wrapping
First, newly-formed 5 uL of DNA was mixed with 0.5uL of 10 ug/mL Ethidium bromide (EtBr, Nacalai Tesque). Fluorescent images were captured using the same set-up as above. A fluorescent filter for GFP (Zeiss) was used for the 200nm beads, and a Cy3.5 filter (ThorLab) was used for EtBr stained DNA.
Fluorescent beads were mixed with the DNA-tile stained with EtBr. The beads were FluoSpheres (carboxylate modified. 0.2 um in diameter, 2 mM surface azide group; Ex/Em= 505/515 nm, Invitrogen). Then, 20 uM at final of Capture DNA was added to wrap the object.
Atomic Force Microscopy (AFM)
The DNA tile solution was spotted in 1uL on mica and the nano-sized structures were observed following the AFM manual for Veeco Manifold Multimode V).